Reproducible research with AnVILPublish
An exploration of elements of reproducible research with the AnVILPublish package. We will illustrate how to make a docker container tailored for publishing AnVIL packages and then emphasize the merits of an R package structure for organizing research activities in a manner that emphasizes provenance and reproducibility.
Learning Objectives
This week we'll explore elements of reproducible research with the AnVILPublish package. We will illustrate how to make a docker container tailored to a particular purpose (in this case, publishing AnVIL packages!). We'll then emphasize the merits of an R package structure for organizing research activities in a manner that emphasizes provenance and reproducibility. The R package structure coupled with git will form the basis of AnVIL workspace creation, allowing us to maintain a single, version-controlled source for Jupyter notebook or RStudio-based AnVIL workspaces.
Key Resources
- Visit https://anvil.terra.bio to use the AnVIL platform.
- The directory and text-file structure of R packages make them easy to write, maintain, and validate; the Writing R Extensions vignette that comes with R is the definitive source; an excellent resource is R Packages (e-)book.
- Docker containers form a basis for reproducibility in AnVIL; we make use of custom docker containers extending the terra-jupyter-bioconductor and anvilproject-rstudio-bioconductor images following instructions at Docker tutorial: Custom cloud environments for Jupyter notebooks (terra-docker/README.md is also useful).
Review
Previously...
- The course schedule contains links and videos of previous sessions
Essential Steps
- Login
- Workspaces
- Billing accounts
- (R-based) Jupyter notebooks or RStudio for interactive analysis
Cloud Computing Environment
- Runtime and persistent disk
- Workspace DATA and buckets
- AnVIL package for interaction with workspace components
Workshop Activities
What's the purpose?
- RStudio provides a rich environment for working in R, but Jupyter notebooks are also relevant, e.g., providing a focused analysis for less-experienced collaborators to walk through.
- We'd like to be able to provide users with documentation that is accessible in either environment.
- The documentation should be consistent across environments.
Provenance is important
- Who wrote or contributed to the software?
- What does the software do?
- What license is it available under?
- What version of the software is currently in use?
Setup
- Log in to AnVIL using the email address you used to register for the course and navigate (via the HAMBURGER) to Workspaces.
- Clone the Bioconductor-Workshop-AnVILPublish workspace
- Unique workspace name
- Billing project: deeppilots-bioconductor-jun7
Start a CUSTOM CLOUD ENVIRONMENT
- 'Cloud Environment' in the top right of the workspace, choose 'Customize'
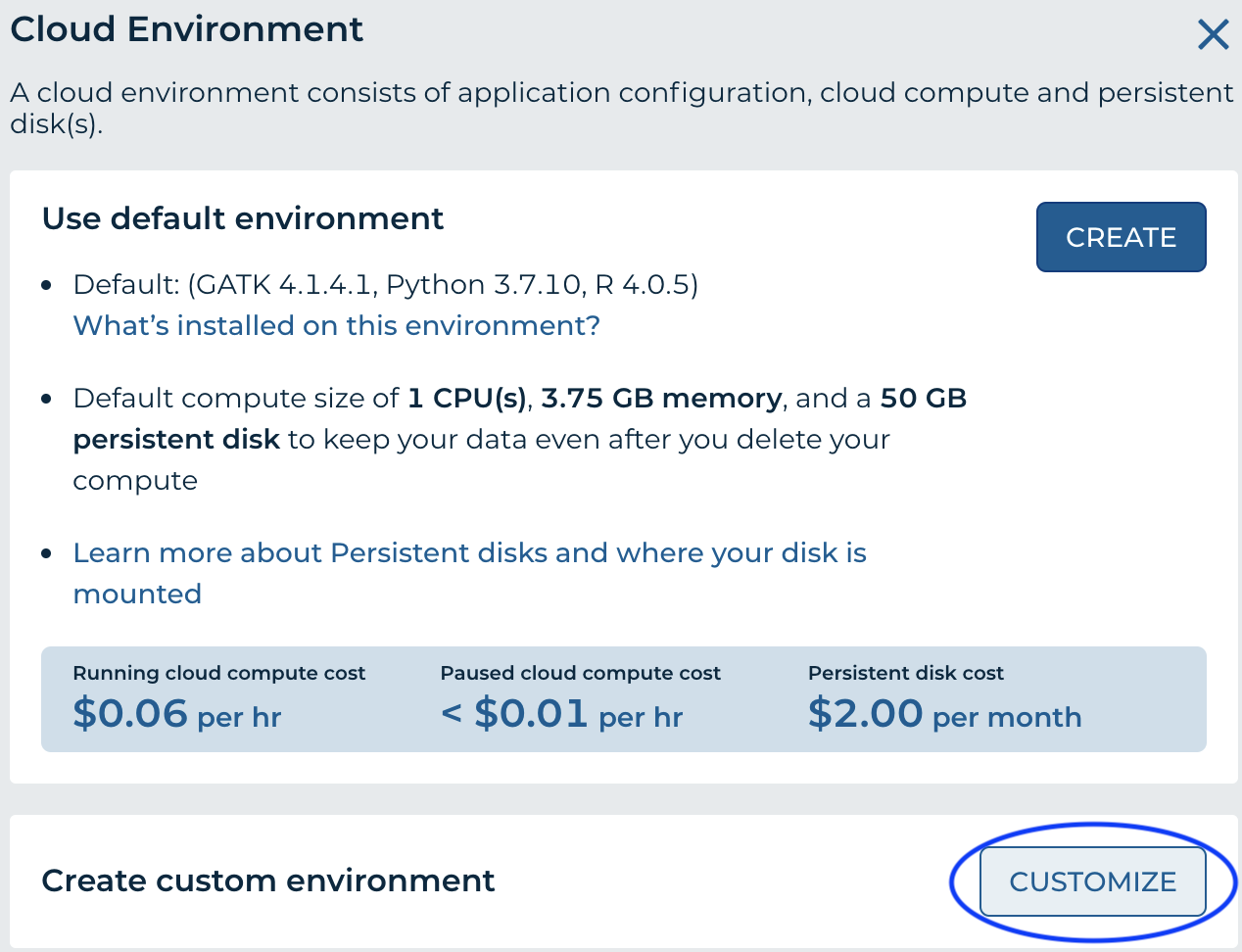
- From the 'Application Configuration' dropdown, choose 'Custom Environment'
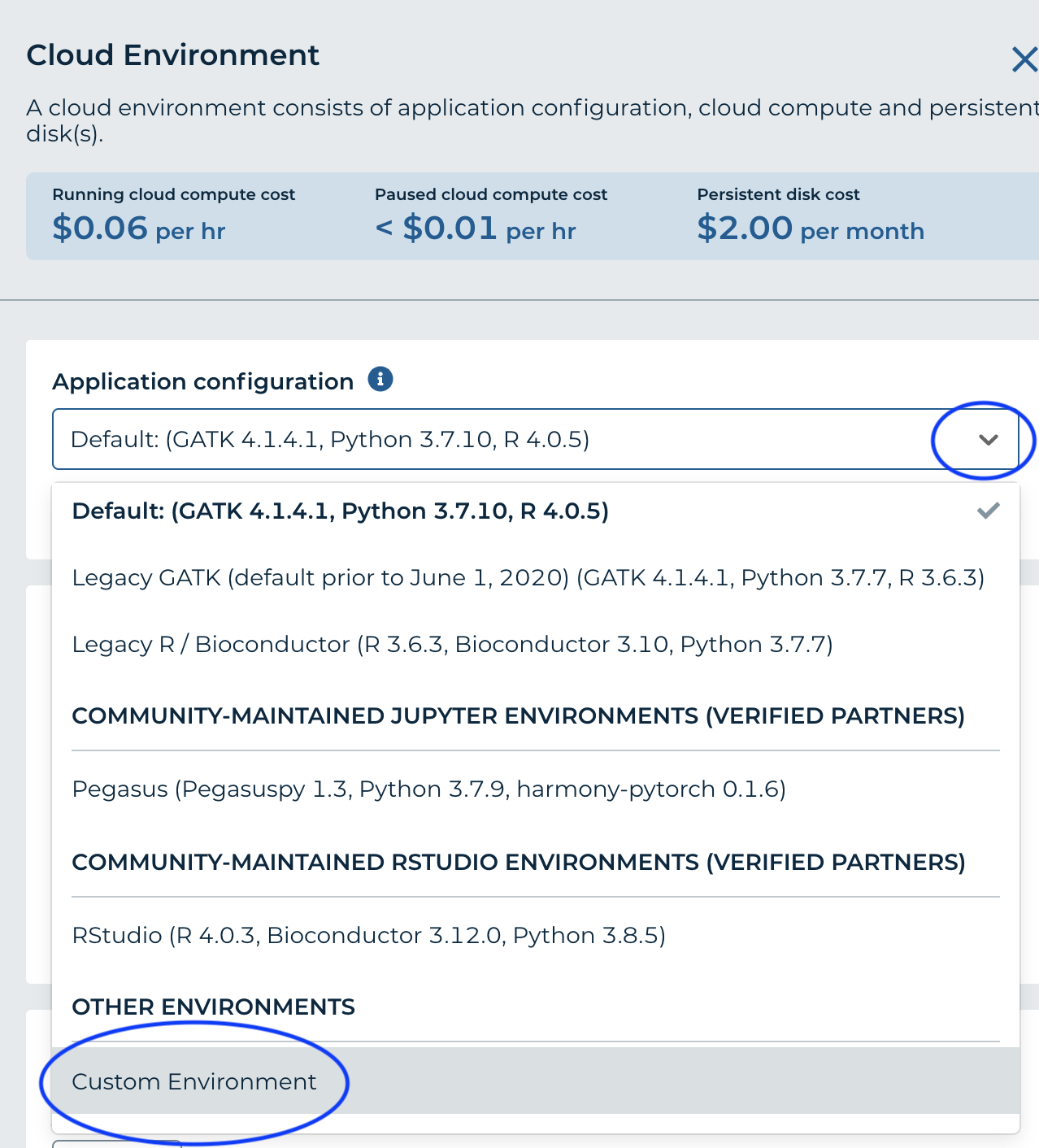
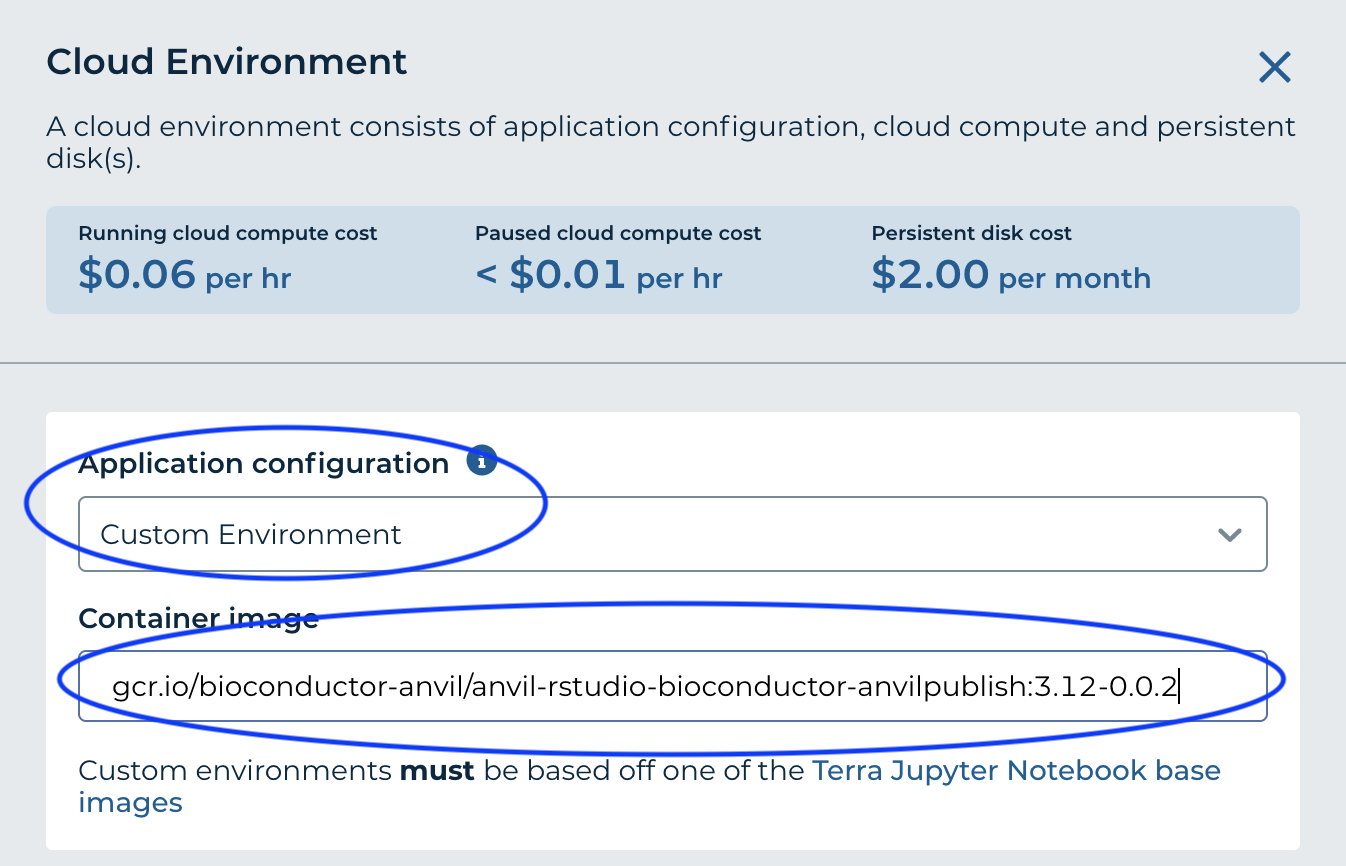
- For 'Container Image' enter gcr.io/bioconductor-anvil/anvil-rstudio-bioconductor-anvilpublish:3.12-0.0.2
R Packages
Create local git clones of the source code of two packages
system2("git", c("clone", "https://github.com/Bioconductor/AnVILPublish"))
system2("git", c("clone", "https://github.com/mtmorgan/AnVILPublishDemo"))
Simple text-based files organize R code, help pages, vignettes, and metadata.
AnVILPublishDemo$ tree
.
├── DESCRIPTION
├── NAMESPACE
├── R
│ └── utilities.R
├── README.md
├── man
│ └── utilities.Rd
└── vignettes
└── A_Introduction.Rmd
Packages are extensible, e.g., all files under an 'inst/' directory are installed with the package
├── inst
│ ├── docker
│ │ ├── anvil-rstudio-bioconductor-anvilpublish
│ │ │ └── Dockerfile
│ │ └── terra-juptyer-bioconductor-anvilpublish
│ │ └── Dockerfile
│ ├── tables
│ │ └── participants.csv
│ └── workflows
│ └── coming_soon.wdl
The DESCRIPTION file provides provenance, including title, version, description, author(s) & their contributions, licensing, as well as system dependencies.
Package: AnVILPublishDemo
Title: Simple Demonstration of AnVILPublish Functionality
Version: 0.0.1
Authors@R:
c(person(
given = "Martin",
family = "Morgan",
role = c("aut", "cre"),
email = "mtmorgan.bioc@gmail.com",
comment = c(ORCID = "0000-0002-5874-8148")
))
Description: AnVILPublish is a way to transform R / Bioconductor
packages, especially vignettes, in Jupyter notebooks for use in
the AnVIL computational environment. The AnVILPublishDemo package
illustrates some of this functionality.
License: Artistic-2.0
Encoding: UTF-8
LazyData: true
Roxygen: list(markdown = TRUE)
RoxygenNote: 7.1.1
Suggests:
knitr,
rmarkdown
VignetteBuilder: knitr
Vignettes
- A natural place to document what the package does in a narrative 'literate programming' manner. If the code in the vignette does not work, then the package does not build and check successfully.
- Vignettes may also contain metadata, e.g., the author and date last revised.
From R Package to AnVIL Workspace
Easy!
AnVILPublish::as_workspace(
"~/AnVILPublishDemo",
"deeppilots-bioconductor-jun7",
"AnVILPublishDemo-YOUR_NAME_HERE",
create = TRUE
)
What do we get?
- DASHBOARD: Provenance -- title, authors, description, version, license
- NOTEBOOKS: ready to evaluate under an R kernel
- DATA: tables from packages added. Interpolation of google bucket possible
- All described in the AnVILPublish vignette
Maybe a little surprising...
- The package can be developed on your own computer (for instance), and published from there, provided gcloud software is installed.
A Little Under the Hood: Custom Docker Files
Summary
What You've Accomplished
Appreciated R package structure
- Organizing components of analysis
- Literate programming
- Provenance
Transforming packages to notebooks and workspaces
- Metadata as DASHBOARD entries, including provenance
- Vignettes as Jupyter notebooks
- DATA tables populated from the package
- Coming soon: addition of workflows; creating workspaces for RStudio
Next Steps
- Follow instructions at Set up billing with $300 Google credits to explore Terra to enable billing for your own projects.
Frequently Asked Question
- What docker images can be used as base images for customization? The main images derive from terra-jupyter-bioconductor (for Jupyter-based images) and anvil-rstudio-bioconductor (for RStudio-based images). Any container can be used in a workflow.
- Can you specify the runtime environment as part of the workspace? This does not seem to be possible at the moment. One could include a notebook or other code that checked the runtime to see that it meets particular conditions, but this would rely on the user running the code.
- Enhance reproducibility by 'fixing' package versions, e.g., using packr? Instead, specify precise package versions in a customized Dockerfile.