What is AnVIL?
The traditional model of genomic data sharing – centralized data warehouses from which researchers download data to analyze locally – is increasingly unsustainable. Not only are transfer/download costs prohibitive, but this approach also leads to redundant siloed compute infrastructure and makes ensuring security and compliance of protected data highly problematic.
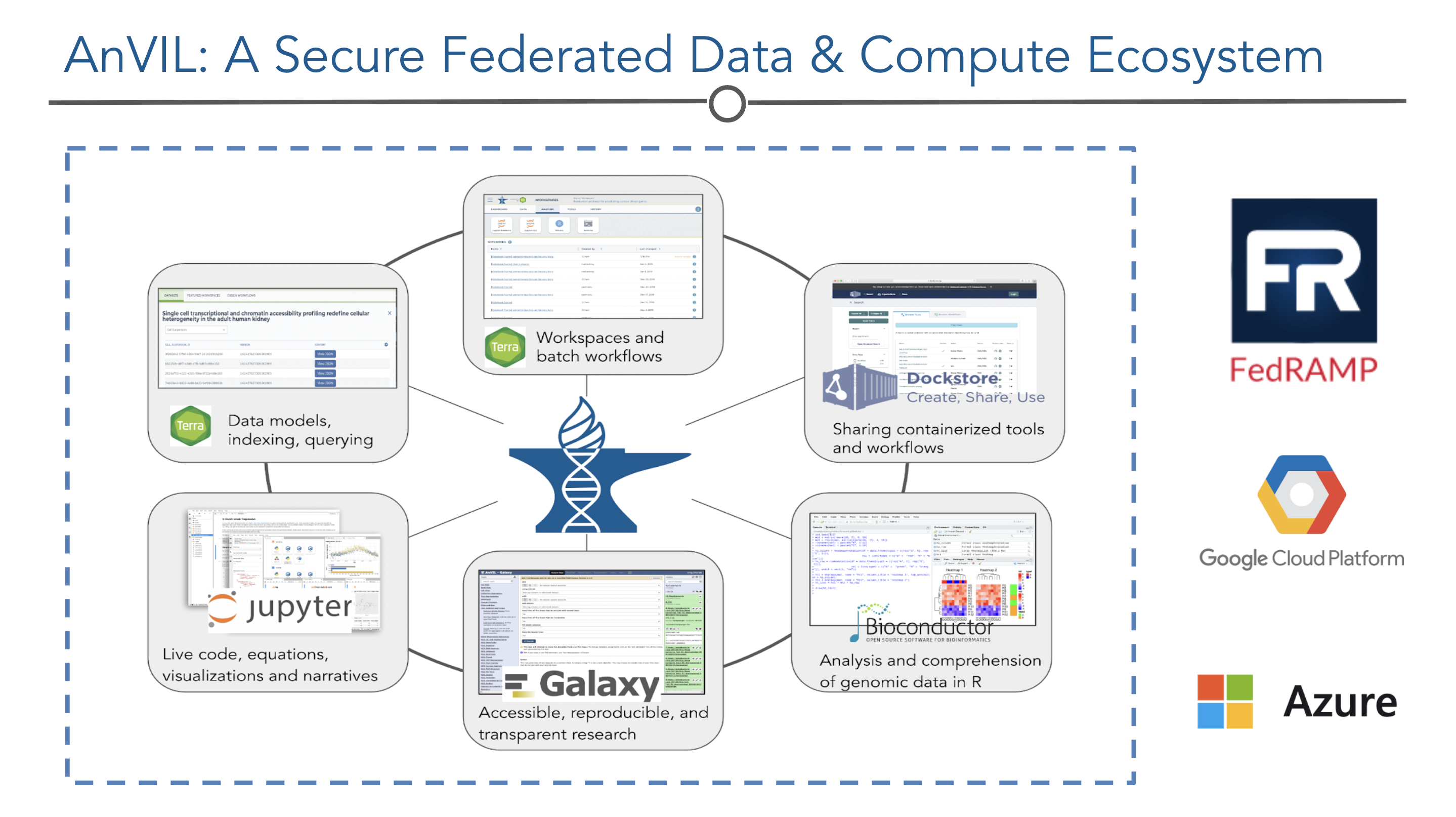
The NHGRI Genomic Data Science Analysis, Visualization, and Informatics Lab-Space, or AnVIL, inverts the traditional model, providing a cloud environment for the analysis of large genomic and related datasets.
By providing a unified environment for data management and compute, AnVIL eliminates the need for data movement, allows for active threat detection and monitoring, and provides elastic, shared computing resources that can be acquired by researchers as needed.
Platform Components
The platform is built on a set of established components that have been used in a number of flagship scientific projects. The Terra platform provides a compute environment with secure data and analysis-sharing capabilities. Dockstore provides standards-based sharing of containerized tools and workflows. The AnVIL Data Explorer allows users to create cross-dataset cohorts over open and managed-access datasets. Bioconductor and Galaxy provide environments for users at different skill levels to construct and execute analyses.
Analysis Tools
AnVIL provides a collaborative environment for creating and sharing data and analysis workflows for both users with limited computational expertise and sophisticated data scientist users.
AnVIL provides multiple entry points for data access and analysis, including execution of batch workflows written in WDL, notebook environments including Jupyter and RStudio, Bioconductor packages for building analysis on top of AnVIL APIs and services, and will offer Galaxy instances for interactive analysis. It will be possible to integrate additional analysis environments through standard APIs.
Datasets
AnVIL provides access to key NHGRI datasets, such as the CCDG (Centers for Common Disease Genomics), CMG (Centers for Mendelian Genomics), eMERGE (Electronic Medical Records and Genomics), as well as other relevant datasets.
Platform Interoperability
AnVIL is a member of the NIH Cloud Platform Interoperability Effort (NCPI) and is collaborating with the NCPI working groups to establish and implement technical standards enabling cross-platform authentication and authorization, cross-platform data discovery, and the cross-platform exchange of datasets, analysis tools, and derived data.
AnVIL is a registered knowledgebase and repository in the FAIRsharing registry of data and metadata standards, inter-related to databases and data policies.
Platform Vision
Long-term, the AnVIL will provide a unified platform for ingestion and organization for a multitude of current and future genomic and genome-related datasets.
Importantly, AnVIL will ease the process of acquiring access to protected datasets for investigators and drastically reduce the burden of performing large-scale integrated analyses across many datasets to fully realize the potential of ongoing data production efforts.
Getting Started
See our Learn section for information on getting started with the AnVIL platform.